3 Promoter sequence sets
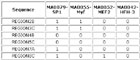
7 ACTB genes and 13 randomly picked genes
7 ACTB genes and 13 randomly picked genes
7 ACTB genes and 13 randomly picked genes
3 Promoter motifs - Sequence logos
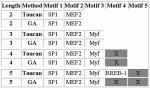
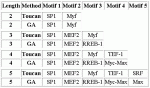
7 ACTB input sequences
7 ACTB input sequences
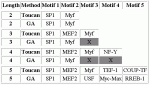
6 ACTB input sequences